Patchwork people
Pharyngula 2024-06-21
Here’s a quick & sloppy science video — I try to explain the ubiquity of mosaicism and how it undermines simplistic ideas of genetic determinism and also, as a bonus, the concept of junk DNA.
Transcript below the fold.
I’m going to tell you a fascinating story, recently published in El Pais, about an interesting genetic condition.
Two eggs fertilized by two sperm coincided in a uterus and, instead of giving rise to two sisters, they fused to form a single person: Karen Keegan. When she was 52 years old, this woman from Boston suffered very serious kidney failure, but luckily she had three children willing to donate a kidney to her. The doctors did genetic tests to see which offspring was most compatible and they got a major surprise: the test said that two of them were not her children. The reality was even more astonishing: Karen Keegan had two different DNA sequences, two genomes, depending on the cell you looked at. Biologist Alfonso Martínez Arias maintains that this chimeric woman is conclusive proof that DNA does not define a person’s identity.
 This is not a surprising event, although I doubt that a medical professional would say that a woman’s children were not her own. I expect a little more discretion! Keegan was a chimera, but not this kind. That is, once upon a time, her mother was carrying a pair of fraternal twins, and at some early point in development, they fused, making a single embryo that carried on. I’ve done this with frogs, if you do it early, when the embryo is only a few hundred cells in size and before any major differentiation has taken place, the cells will simply mingle together. I’ve also done the experiment with sea urchin blastulae — if done before polarity is set up, they’ll just happily fuse to make an embryo with cells from two different sources.
This is not a surprising event, although I doubt that a medical professional would say that a woman’s children were not her own. I expect a little more discretion! Keegan was a chimera, but not this kind. That is, once upon a time, her mother was carrying a pair of fraternal twins, and at some early point in development, they fused, making a single embryo that carried on. I’ve done this with frogs, if you do it early, when the embryo is only a few hundred cells in size and before any major differentiation has taken place, the cells will simply mingle together. I’ve also done the experiment with sea urchin blastulae — if done before polarity is set up, they’ll just happily fuse to make an embryo with cells from two different sources.
It differs from conjoined twins, which is what you get when the fusion takes place later, after major organs and their organization has been determined. Chimerism can be practically undetectable in the absence of obvious allelic differences or genetic tests.
The surprising conclusion, to some people at least, is that this implies that DNA does not define a person’s identity. I agree. Genetics provides the raw material, but the environment and circumstances of development play a far greater part in that mysterious phenomenon called “identity”. A key feature of our bodies is plasticity — the genome generates a multi-functional state machine that is shaped by our life experiences.
However, I don’t think chimerism is the best evidence that genes and phenotype are to some degree disconnected. I think the better indicator is the prevalence of mosaicism, which is far more common.
What’s the difference? Chimerism is the result of fusing two initially separate individuals; mosaicism is the result of cells within a lineage accumulating mutations or epigenetic differences that make them distinct. Mosaicism is extremely common, although it varies in degree. I would estimate that only a few individuals in tens of thousands are chimeric, but that everyone reading this right now is mosaic. It’s an inevitable consequence of errors in cell division that not all the products of mitosis are going to be genetically identical.
 Chimerism often has minimal, even negligible effects — we can tolerate minor variations in the genomic sequence. However, it can also have huge, life-threatening effects. Cancer, for instance, is a patch of tissue that has acquired variations that differ from the original. There are even more unusual consequences, like this report of identical twins, one of which had Down syndrome. Just one. They were genetically identical, but one had Trisomy 21. Everything was the same except one whole chromosome was duplicated in one individual.
Chimerism often has minimal, even negligible effects — we can tolerate minor variations in the genomic sequence. However, it can also have huge, life-threatening effects. Cancer, for instance, is a patch of tissue that has acquired variations that differ from the original. There are even more unusual consequences, like this report of identical twins, one of which had Down syndrome. Just one. They were genetically identical, but one had Trisomy 21. Everything was the same except one whole chromosome was duplicated in one individual.
How can that happen? I’ll remind you that every single division of every cell in your body has the potential for error — there is an error rate for mutational changes in the DNA sequence, and also for mistakes in the segregation of chromosomes during mitosis. These are very low frequencies, but given that you are made up of 30 trillion cells, each of which is the product of multiple cell divisions, the likelihood of multiple mutant clones of tissue in your body approaches certainty.
To explain how we could get identical twins with a rather profound genetic difference, I have to explain and review the basic process of mitotic cell division. You’ve probably heard about this before, way back in grade school possibly, so this is a quick reminder.
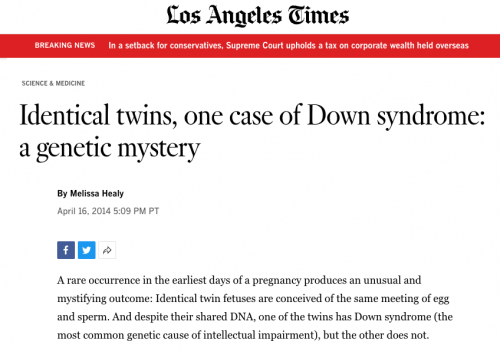
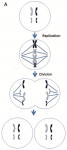 Every somatic cell has paired chromosomes. We have 23 pairs of chromosomes, but this diagram illustrates a cell with just 2 pairs, or a total of 4 chromosomes, to keep it simple. In mitosis, the first step is replication — the DNA of each chromosome is replicated, but the two copies of DNA remain associated at the centromere. The two copies are pulled apart and move to opposite poles of the cell during division, and then the cell membranes separate. The end result is two cells with identical DNA organized in identical chromosomes. These cells are called euploid, meaning they have an integral multiple of the number of chromosomes found in the egg or sperm. The alternative is to be aneuploid, that is to have a deviation in the appropriate number. That can happen fairly easily by a couple of different mechanisms.
Every somatic cell has paired chromosomes. We have 23 pairs of chromosomes, but this diagram illustrates a cell with just 2 pairs, or a total of 4 chromosomes, to keep it simple. In mitosis, the first step is replication — the DNA of each chromosome is replicated, but the two copies of DNA remain associated at the centromere. The two copies are pulled apart and move to opposite poles of the cell during division, and then the cell membranes separate. The end result is two cells with identical DNA organized in identical chromosomes. These cells are called euploid, meaning they have an integral multiple of the number of chromosomes found in the egg or sperm. The alternative is to be aneuploid, that is to have a deviation in the appropriate number. That can happen fairly easily by a couple of different mechanisms.
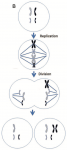 One mechanism is nondisjunction. This is an error in which the duplicated DNA are NOT pulled apart, so two identical chromosomes are pulled away from one daughter cell and end up in the other daughter cell. The end result: two aneuploid cells, one lacking a chromosome, the other having an extra chromosome. If this had been Chromosome 21, for instance, the cell on the right would be trisomic and would have the Down syndrome trait. (Trisomic means having 3 copies of a chromosome, rather than the typical disomy, having two copies. Monosomy is having only one copy.)
One mechanism is nondisjunction. This is an error in which the duplicated DNA are NOT pulled apart, so two identical chromosomes are pulled away from one daughter cell and end up in the other daughter cell. The end result: two aneuploid cells, one lacking a chromosome, the other having an extra chromosome. If this had been Chromosome 21, for instance, the cell on the right would be trisomic and would have the Down syndrome trait. (Trisomic means having 3 copies of a chromosome, rather than the typical disomy, having two copies. Monosomy is having only one copy.)
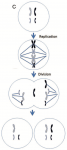 There are other ways to produce aneuploid cells during mitotic divisions. One is anaphase lagging — the chromosomes separate properly, but one has lost it’s connection to the fibers that pull the chromosomes about, and basically can be lost. Another process is called endoreplication, in which a chromosome is duplicated an additional time. If this kind of error occurred at the first division of an embryo, half the cells would be trisomic; the later the cell division, the smaller the proportion of cells carrying the error.
There are other ways to produce aneuploid cells during mitotic divisions. One is anaphase lagging — the chromosomes separate properly, but one has lost it’s connection to the fibers that pull the chromosomes about, and basically can be lost. Another process is called endoreplication, in which a chromosome is duplicated an additional time. If this kind of error occurred at the first division of an embryo, half the cells would be trisomic; the later the cell division, the smaller the proportion of cells carrying the error.
Here’s an example. One of the cells in a human embryo undergoes a mitotic non-disjunction during the early cleavage stages, so a handful of cells have the normal complement of 46 chromosomes, one has 47, and another has 45 chromosomes. These cells proliferate to produce a trophectoderm (which will form the placenta) and an inner cell mass (which will make up the embryo) that are a mosaic of cells with 46 and 47 chromosomes. If a twinning event occurred, you could end up with two individuals, one with Down syndrome and the other without. 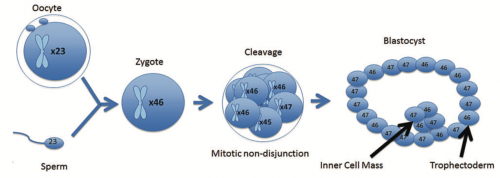
You might be wondering how often this happens, not just mosaicism for Trisomy 21, but all kinds of variations in the genome. It’s not clear. One reason is that aneuploid cells do not replicate as well as euploid cells, so the proportion can vary over time — they can be swamped out by healthy cells. Monosomic cells in particular may be lacking essential genes, and can simply die off — the cell with 45 chromosomes in this diagram disappeared. Trisomic cells may also spontaneously lose the extra chromosome by anaphase lagging, returning the cell to the euploid state (although that introduces new complications — I’ll mention uniparental disomy in a moment). It’s also difficult to assess because…how do you assess the genetic state of each cell in an embryonic mass when there’s no obvious morphological difference?
One review paper says “Mosaicism occurs in 15–90% of all cleavage stage human embryos”. 15-90%? That’s a huge spread. The problem is sampling — especially with human embryos, you can’t often do the thorough assessment of dismantling the embryo and doing a chromosome analysis of each and every cell.
Another paper by van Echten-Arends found that 73% of cleavage stage embryos were mosaic, but 59% of all later embryos were mosaic. The point is that this is a common event, but profoundly damaged cells tend to be gradually lost as the embryo gets older. In prenatal examinations, only 1 or 2% of fetuses exhibit detectable mosaicism, a consequence of loss of damaged cells, the greater replication rate of healthy cells, and the impossibility of doing exhaustive analyses of every cell in a fetus. These are also fairly coarse cytological measures of raw chromosome numbers — it is practically certain that everyone is mosaic for smaller differences in their genetic sequences. Keep in mind that every infant is born with 100-150 mutations inherited from meiotic errors in their parents’ gametes, and those kinds of errors don’t stop at fertilization. Even some of the chromosomal variants are effectively invisible!
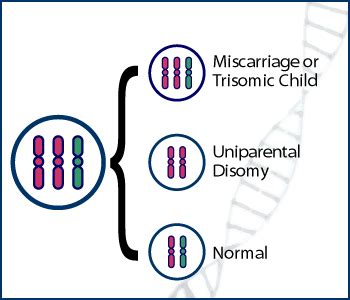 For example, say you’ve got a cell that is trisomic, that is, it carries two copies of a paternal chromosome and one copy of a maternal chromosome. Cells can shed one of those three chromosomes, but which one? If it loses one of the paternal chromosomes, the cell is now euploid, disomic, and carries one paternal and one maternal chromosome, the normal state of affairs, and will develop unexceptionally. If it loses the maternal chromosome, though, it’s euploid and disomic, but now has two paternal chromosomes, which you might think wouldn’t matter…but chromosomes in sperm are imprinted, that is, marked with a different pattern of histones, than are chromosomes in the ova. We don’t know enough about the effects of imprinting to say for sure, but there may be interesting differences in behavior between cells with exclusively paternal or maternal imprinting from cells with a balance of paternal and maternal imprinting. In some cases it’s not just what genes you inherit, but whether you got them from your mother or your father, and in some cases of uniparental disomy you may have patches of tissue that are exclusively carrying genes modified by only your mother’s or your father’s gene regulation configuration.
For example, say you’ve got a cell that is trisomic, that is, it carries two copies of a paternal chromosome and one copy of a maternal chromosome. Cells can shed one of those three chromosomes, but which one? If it loses one of the paternal chromosomes, the cell is now euploid, disomic, and carries one paternal and one maternal chromosome, the normal state of affairs, and will develop unexceptionally. If it loses the maternal chromosome, though, it’s euploid and disomic, but now has two paternal chromosomes, which you might think wouldn’t matter…but chromosomes in sperm are imprinted, that is, marked with a different pattern of histones, than are chromosomes in the ova. We don’t know enough about the effects of imprinting to say for sure, but there may be interesting differences in behavior between cells with exclusively paternal or maternal imprinting from cells with a balance of paternal and maternal imprinting. In some cases it’s not just what genes you inherit, but whether you got them from your mother or your father, and in some cases of uniparental disomy you may have patches of tissue that are exclusively carrying genes modified by only your mother’s or your father’s gene regulation configuration.
Yeah, it gets complicated, I know. And that diagram shows a single cell — you’ve almost certainly got patches of tissue in various parts of your body that vary from the initial genetic pattern established at fertilization. It is entirely possible to have a brain, or parts of a brain, that have a different genetic combination or even different chromosome arrangement from the rest of your body. Sometimes people talk about how their brain doesn’t match their body, and this is physically, genetically possible (although I’d suggest that environment and culture and upbringing are probably a greater contributor to that feeling than somatic mosaicism…but who knows? That would be a difficult difference to measure given current technology.)
But that’s not all. Cells divide throughout your lifetime, and as you get older, there is increasing divergence from the initial sequence. 20% of men over 60 (hey, that includes me) have white blood cells that show loss of the Y chromosome — they’re aneuploid!
Mosaic loss of chromosome Y (LOY) in circulating white blood cells is the most common form of clonal mosaicism yet our knowledge of the causes and consequences of this is limited. Here, using a computational approach, we estimate that 20% of the male population represented in the UK Biobank study (n = 205,011) has detectable LOY. … We demonstrate that genetic susceptibility to LOY is associated with non-haematological effects on health in both men and women, which supports the hypothesis that clonal haematopoiesis is a biomarker of genomic instability in other tissues. … Collectively, these data highlight the value of studying clonal mosaicism to uncover fundamental mechanisms that underlie cancer and other ageing-related diseases.
The key phrase here is that this number is a marker of genomic instability. They only looked at one class of cells for one kind of chromosome defect, and it’s surprisingly high. But there are other tissues and other chromosomes that could be examined, and they all would show evidence of increasing mosaicism with age. We’re all gradually changing at a genetic level as we get older.
There’s another level of divergence, too: epigenetic drift. Our chromosomes are all packaged up in proteins, histones, that play a significant role in gene regulation: if histones are methylated, they pack the DNA more tightly, limiting the access of enzymes and downregulating expression; if the histones are acetylated, the DNA sprawls open and genes within that region are more readily activated. 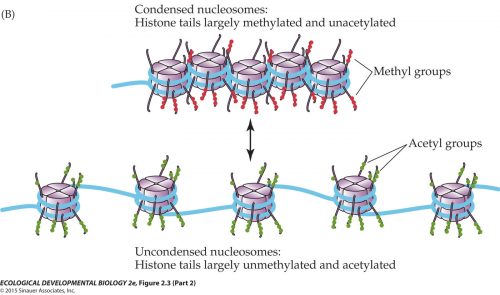
Unsurprisingly, methylation patterns vary in different tissus. You might expect that, since different tissues have different patterns of gene activity. What’s interesting is that these patterns change with age, and that the degree of methylation is a clocklike marker of chronological age. Here we can see that the methylation age determined from cells in saliva, blue, (primarily epithelial cells) tracks the chronological age fairly closely, same as age from methylation of DNA in blood, in red. One analysis claims to be able to predict someone’s age within plus or minus two years from methylation state, although the data shown in these graphs makes that seem like unlikely precision, to me. Still, those are pretty good correlations. 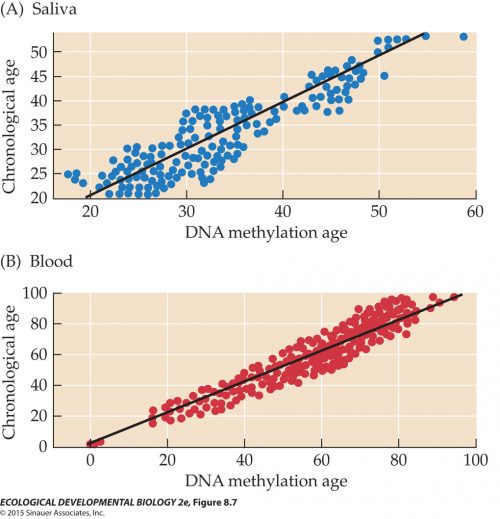
I should also mention that all this changing methylation, which is roughly equivalent to increasing inactivation over time, is another data point in the idea that most DNA is junk. We can methylate an awful lot of our genome without significant deleterious effect, because most of that DNA is already non-functional.
Twin studies also show the trend in aging. Here we see a comparison of the 5 methyl cytosine compared between pairs of 3 year old twins, and it’s low. Looking at 50 year old twins, it’s greatly increased, and further, the twins have greater disparity to each other. Similarly with acetylation of histone H3 (remember, acetylation correlates with activation), and you can see that it’s very similar and relatively high in pairs of 3 year old twins, and it declines in the 50 year old twins and they also become more different from each other. 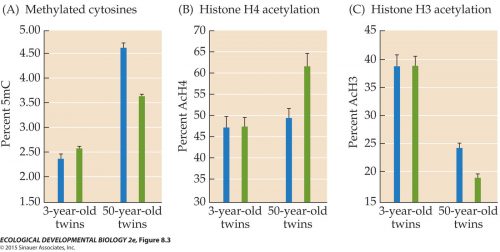 We can get more detailed and look at specific bands in chromosomes. This figure uses a novel technique of generating probes that label specific methylation sites in chromosomes. For one twin of a pair, you make a red probe to label wherever Twin A has methylated their chromosome; then you make a similar set of green probes for wherever Twin B has methylation. Then you apply both probes to a set of chromosomes. Wherever both Twin A and Twin B have methylation, you’ll get both red and green fluorescence, so it’ll glow yellow. Look at the chromosomes from 3 year old twins; they’re all yellow, because they start out with nearly identical methylation patterns.
We can get more detailed and look at specific bands in chromosomes. This figure uses a novel technique of generating probes that label specific methylation sites in chromosomes. For one twin of a pair, you make a red probe to label wherever Twin A has methylated their chromosome; then you make a similar set of green probes for wherever Twin B has methylation. Then you apply both probes to a set of chromosomes. Wherever both Twin A and Twin B have methylation, you’ll get both red and green fluorescence, so it’ll glow yellow. Look at the chromosomes from 3 year old twins; they’re all yellow, because they start out with nearly identical methylation patterns. 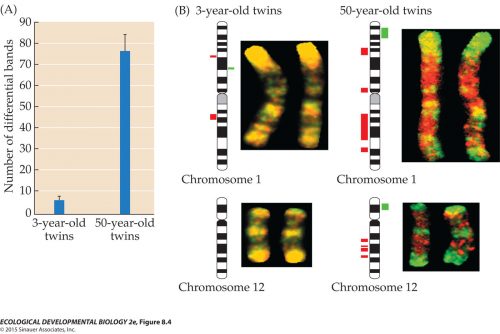
Then look at the 50 year old twins. Wherever it’s green, that says only Twin B methylated this region. Wherever it’s red, only Twin A methylated that spot. This is called epigenetic drift, where the pattern of epigenetic marks gradually changes over time, and not in a deterministic way. Your brother or sister, even if they are a monozygotic twin, is steadily diverging away from you over time, partly by chance, and partly as a consequence of different environmental circumstances. Most of these changes are inconsequential, since much of the genome is junk; some will be a cause of the symptoms of aging, or may lead to cancers.
To return to Alfonso Martínez Arias’s claim that DNA does not define a person’s identity, I agree. That’s trivially true. DNA changes over time, it is modified differently in different tissues, it is extensively modified by chance and by the environment, some of us are chimeras and all of us are mosaic. Our genomes constitute a baseline from which individual cells vary in unpredictable ways. We are all of us patchwork people, because our DNA is not constant and not uniform, within limits. We cannot be defined by only our DNA, because we have to take into account all the modifications of DNA that take place over the course of development.
And of course, brains are even more variable and affected by day to day, even hour to hour or second to second events, and you cannot derive a mind by computing the genome.
You know, I teach a classical transmission genetics course, and I rarely have time to squeeze in this kind of information in there. I got to teach an ecological developmental biology course this past term where I finally got to let my freak flag fly, and it was so much fun — I hope to do it again in two years. You can find much of this information in the textbook for that course, Ecological Development, by Gilbert and Epel. But also, if you read much molecular genetics, this is all common knowledge — I’ll include citations of a couple of papers below.
The fusion of two sisters into a single woman suggests that human identity is not in our DNA
Identical twins, one case of Down syndrome.
Jeremy Thorpe, Ikeoluwa A. Osei-Owusu, Bracha Erlanger Avigdor, Rossella Tuplerl, Jonathan Pevsner (2020) Mosaicism in Human Health and Disease. Annu. Rev. Genet. 2020. 54:487–510.
Vijg J (2000) Somatic mutations and aging: a re-evaluation. Mutation Research 447: 117–135.
Thompson DJ et al. (2019) Genetic predisposition to mosaic Y chromosome loss in blood. Nature 575:652-657,